Protein C-ETS2 is a protein that in humans is encoded by the ETS2 gene. The protein encoded by this gene belongs to the ETS family of transcription factors. It has been found to play a key role in the development of inflammatory bowel disease.
Interactions
ETS2 has been shown to interact with:
- C-jun,
- Cyclin-dependent kinase 10,
- ERG,
- myb, and
- ZMYND11.
Clinical significance
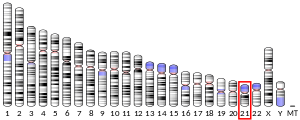
ETS2 has been linked to a variety of diseases, including Takayasu's arteritis, inflammatory bowel disease, primary sclerosing cholangitis, and ankylosing spondylitis, all associated with a region on chromosome 21 at 21q22.
References
- ^ GRCh38: Ensembl release 89: ENSG00000157557 – Ensembl, May 2017
- ^ GRCm38: Ensembl release 89: ENSMUSG00000022895 – Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Dwyer J, Li H, Xu D, Liu JP (Oct 2007). "Transcriptional regulation of telomerase activity: roles of the Ets transcription factor family". Annals of the New York Academy of Sciences. 1114 (1): 36–47. Bibcode:2007NYASA1114...36D. doi:10.1196/annals.1396.022. PMID 17986575. S2CID 44532648.
- "Entrez Gene: ETS2 V-ets erythroblastosis virus E26 oncogene homolog 2 (avian)".
- Chen Y, Ying Y, Wang M, Ma C, Jia M, Shi L, et al. (January 2023). "A distal super-enhancer activates oncogenic ETS2 via recruiting MECOM in inflammatory bowel disease and colorectal cancer". Cell Death & Disease. 14 (1): 8. doi:10.1038/s41419-022-05513-1. PMC 9822945. PMID 36609474.
- Stankey CT, Bourges C, Haag LM, Turner-Stokes T, Piedade AP, Palmer-Jones C, et al. (June 2024). "A disease-associated gene desert directs macrophage inflammation through ETS2". Nature. 630 (8016): 447–456. Bibcode:2024Natur.630..447S. doi:10.1038/s41586-024-07501-1. PMC 11168933. PMID 38839969.
- ^ Basuyaux JP, Ferreira E, Stéhelin D, Butticè G (October 1997). "The Ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner". The Journal of Biological Chemistry. 272 (42): 26188–26195. doi:10.1074/jbc.272.42.26188. PMID 9334186.
- Kasten M, Giordano A (April 2001). "Cdk10, a Cdc2-related kinase, associates with the Ets2 transcription factor and modulates its transactivation activity". Oncogene. 20 (15): 1832–1838. doi:10.1038/sj.onc.1204295. PMID 11313931.
- Dudek H, Tantravahi RV, Rao VN, Reddy ES, Reddy EP (February 1992). "Myb and Ets proteins cooperate in transcriptional activation of the mim-1 promoter". Proceedings of the National Academy of Sciences of the United States of America. 89 (4): 1291–1295. Bibcode:1992PNAS...89.1291D. doi:10.1073/pnas.89.4.1291. PMC 48435. PMID 1741383.
- Wei G, Schaffner AE, Baker KM, Mansky KC, Ostrowski MC (2003). "Ets-2 interacts with co-repressor BS69 to repress target gene expression". Anticancer Research. 23 (3A): 2173–2178. PMID 12894593.
- Stankey CT, Bourges C, Haag LM, Turner-Stokes T, Piedade AP, Palmer-Jones C, et al. (June 2024). "A disease-associated gene desert directs macrophage inflammation through ETS2". Nature. 630 (8016): 447–456. Bibcode:2024Natur.630..447S. doi:10.1038/s41586-024-07501-1. PMC 11168933. PMID 38839969.
Further reading
- Boulukos KE, Pognonec P, Sariban E, Bailly M, Lagrou C, Ghysdael J (March 1990). "Rapid and transient expression of Ets2 in mature macrophages following stimulation with cMGF, LPS, and PKC activators". Genes & Development. 4 (3): 401–409. doi:10.1101/gad.4.3.401. PMID 2186967.
- Watson DK, Mavrothalassitis GJ, Jorcyk CL, Smyth FE, Papas TS (October 1990). "Molecular organization and differential polyadenylation sites of the human ETS2 gene". Oncogene. 5 (10): 1521–1527. PMID 2250910.
- Watson DK, McWilliams MJ, Lapis P, Lautenberger JA, Schweinfest CW, Papas TS (November 1988). "Mammalian ets-1 and ets-2 genes encode highly conserved proteins". Proceedings of the National Academy of Sciences of the United States of America. 85 (21): 7862–7866. Bibcode:1988PNAS...85.7862W. doi:10.1073/pnas.85.21.7862. PMC 282297. PMID 2847145.
- Watson DK, McWilliams-Smith MJ, Nunn MF, Duesberg PH, O'Brien SJ, Papas TS (November 1985). "The ets sequence from the transforming gene of avian erythroblastosis virus, E26, has unique domains on human chromosomes 11 and 21: both loci are transcriptionally active". Proceedings of the National Academy of Sciences of the United States of America. 82 (21): 7294–7298. Bibcode:1985PNAS...82.7294W. doi:10.1073/pnas.82.21.7294. PMC 391330. PMID 2997781.
- Le Beau MM, Rowley JD, Sacchi N, Watson DK, Papas TS, Diaz MO (November 1986). "Hu-ets-2 is translocated to chromosome 8 in the t(8;21) in acute myelogenous leukemia". Cancer Genetics and Cytogenetics. 23 (3): 269–274. doi:10.1016/0165-4608(86)90189-5. PMID 3021321.
- Fujiwara S, Fisher RJ, Seth A, Bhat NK, Showalter SD, Zweig M, et al. (February 1988). "Characterization and localization of the products of the human homologs of the v-ets oncogene". Oncogene. 2 (2): 99–103. PMID 3285299.
- Sumarsono SH, Wilson TJ, Tymms MJ, Venter DJ, Corrick CM, Kola R, et al. (February 1996). "Down's syndrome-like skeletal abnormalities in Ets2 transgenic mice". Nature. 379 (6565): 534–537. Bibcode:1996Natur.379..534H. doi:10.1038/379534a0. PMID 8596630. S2CID 4365956.
- Basuyaux JP, Ferreira E, Stéhelin D, Butticè G (October 1997). "The Ets transcription factors interact with each other and with the c-Fos/c-Jun complex via distinct protein domains in a DNA-dependent and -independent manner". The Journal of Biological Chemistry. 272 (42): 26188–26195. doi:10.1074/jbc.272.42.26188. PMID 9334186.
- Blumenthal SG, Aichele G, Wirth T, Czernilofsky AP, Nordheim A, Dittmer J (April 1999). "Regulation of the human interleukin-5 promoter by Ets transcription factors. Ets1 and Ets2, but not Elf-1, cooperate with GATA3 and HTLV-I Tax1". The Journal of Biological Chemistry. 274 (18): 12910–12916. doi:10.1074/jbc.274.18.12910. PMID 10212281.
- Jayaraman G, Srinivas R, Duggan C, Ferreira E, Swaminathan S, Somasundaram K, et al. (June 1999). "p300/cAMP-responsive element-binding protein interactions with ets-1 and ets-2 in the transcriptional activation of the human stromelysin promoter". The Journal of Biological Chemistry. 274 (24): 17342–17352. doi:10.1074/jbc.274.24.17342. PMID 10358095.
- Kilpatrick LM, Kola I, Salamonsen LA (July 1999). "Transcription factors Ets1, Ets2, and Elf1 exhibit differential localization in human endometrium across the menstrual cycle and alternate isoforms in cultured endometrial cells". Biology of Reproduction. 61 (1): 120–126. doi:10.1095/biolreprod61.1.120. PMID 10377039.
- Rameil P, Lécine P, Ghysdael J, Gouilleux F, Kahn-Perlès B, Imbert J (April 2000). "IL-2 and long-term T cell activation induce physical and functional interaction between STAT5 and ETS transcription factors in human T cells". Oncogene. 19 (17): 2086–2097. doi:10.1038/sj.onc.1203542. PMID 10815800.
- Park US, Park SK, Lee YI, Park JG, Lee YI (July 2000). "Hepatitis B virus-X protein upregulates the expression of p21waf1/cip1 and prolongs G1-->S transition via a p53-independent pathway in human hepatoma cells". Oncogene. 19 (30): 3384–3394. doi:10.1038/sj.onc.1203674. PMID 10918595.
- Pastorcic M, Das HK (November 2000). "Regulation of transcription of the human presenilin-1 gene by ets transcription factors and the p53 protooncogene". The Journal of Biological Chemistry. 275 (45): 34938–34945. doi:10.1074/jbc.M005411200. PMID 10942770.
- Smith JL, Schaffner AE, Hofmeister JK, Hartman M, Wei G, Forsthoefel D, et al. (November 2000). "ets-2 is a target for an akt (Protein kinase B)/jun N-terminal kinase signaling pathway in macrophages of motheaten-viable mutant mice". Molecular and Cellular Biology. 20 (21): 8026–8034. doi:10.1128/MCB.20.21.8026-8034.2000. PMC 86413. PMID 11027273.
- Mullick J, Anandatheerthavarada HK, Amuthan G, Bhagwat SV, Biswas G, Camasamudram V, et al. (May 2001). "Physical interaction and functional synergy between glucocorticoid receptor and Ets2 proteins for transcription activation of the rat cytochrome P-450c27 promoter". The Journal of Biological Chemistry. 276 (21): 18007–18017. doi:10.1074/jbc.M100671200. PMID 11279115.
- Kasten M, Giordano A (April 2001). "Cdk10, a Cdc2-related kinase, associates with the Ets2 transcription factor and modulates its transactivation activity". Oncogene. 20 (15): 1832–1838. doi:10.1038/sj.onc.1204295. PMID 11313931.
- Ezashi T, Ghosh D, Roberts RM (December 2001). "Repression of Ets-2-induced transactivation of the tau interferon promoter by Oct-4". Molecular and Cellular Biology. 21 (23): 7883–7891. doi:10.1128/MCB.21.23.7883-7891.2001. PMC 99954. PMID 11689681.
- Xu XR, Huang J, Xu ZG, Qian BZ, Zhu ZD, Yan Q, et al. (December 2001). "Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver". Proceedings of the National Academy of Sciences of the United States of America. 98 (26): 15089–15094. Bibcode:2001PNAS...9815089X. doi:10.1073/pnas.241522398. PMC 64988. PMID 11752456.
- Islas JF, Liu Y, Weng KC, Robertson MJ, Zhang S, Prejusa A, et al. (August 2012). "Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors". Proceedings of the National Academy of Sciences of the United States of America. 109 (32): 13016–13021. Bibcode:2012PNAS..10913016I. doi:10.1073/pnas.1120299109. PMC 3420197. PMID 22826236.
External links
| PDB gallery | |
|---|---|
| Transcription factors and intracellular receptors | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| see also transcription factor/coregulator deficiencies | |||||||||||||||||||||||||||||||